Many functions of the gastrointestinal tract (GIT) microbiome have been identified. However research indicates that what is currently known in this area is just the tip of the iceberg, with science continually making new discoveries. One such area of discovery is the complex, two-way interaction between exogenous and endogenous toxicity and the GIT microbiome.
Toxicity, in the context of human health, can be considered to be exposure to exogenous and endogenous substances that have the ability to adversely influence physiological homeostasis. Sources of exogenous toxicity, often referred to as xenobiotics, includes pharmaceutical medications, tobacco smoke, alcohol, pesticides, environmental pollutants and endocrine-disrupting chemicals.[1,2]
The metabolism of such xenobiotics can also be a significant source of endogenous toxicity, particularly when hepatic and GIT functionality is suboptimal.
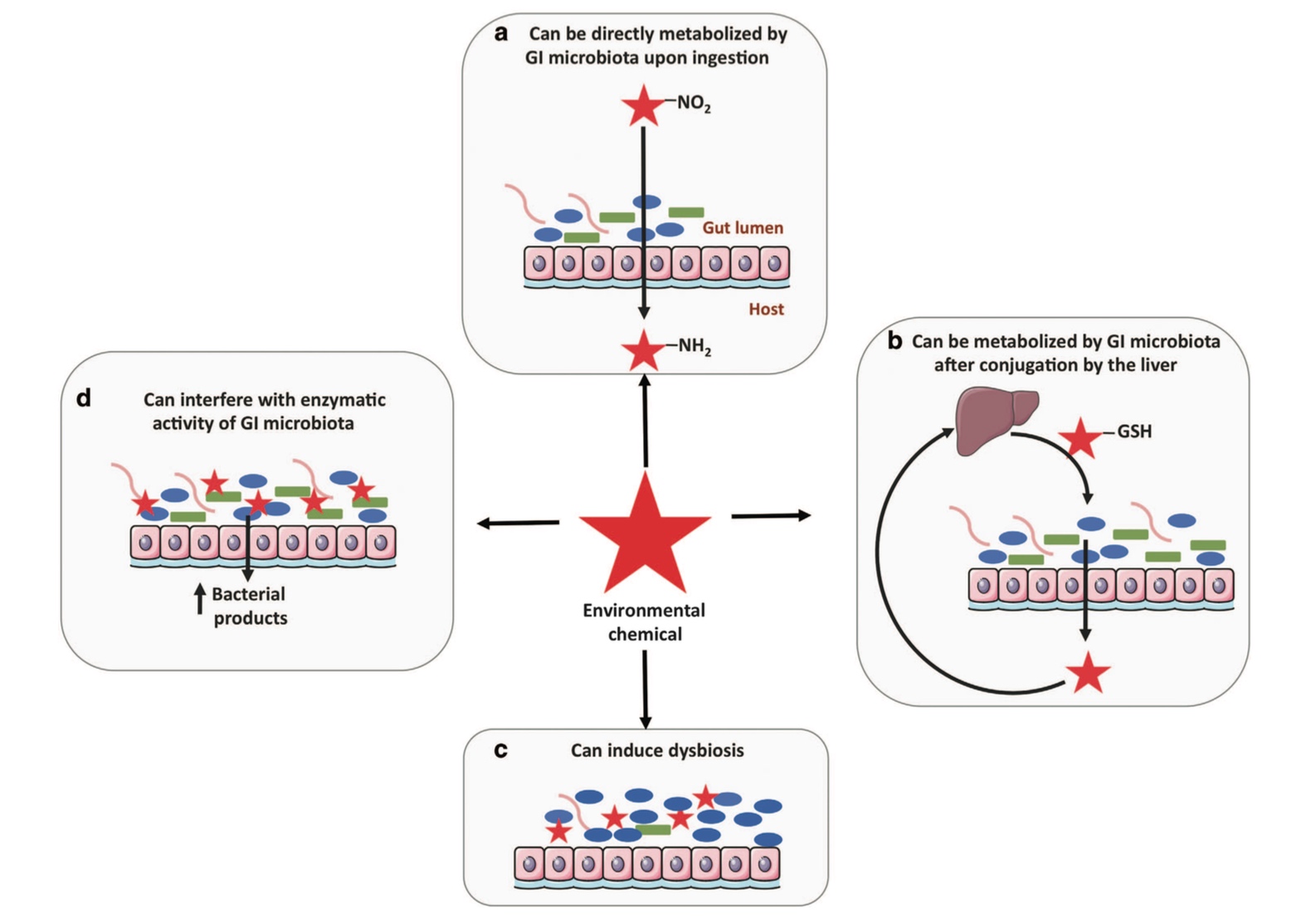
Image Source [6]
The metabolism of xenobiotics involves both local and interdependent processes occurring in the liver and GIT microbiome.[3] Just as the extent of toxicity exposure varies between individuals, so too does the physiological impact of such toxins and our specific capacity to metabolise them. This is significantly influenced by the composition and consequent functionality of the GIT microbiome, which plays an essential role in xenobiotic metabolism and the degree to which it occurs within individuals,[3-6] and whether a xenobiotic substance is reabsorbed, reconverted back to its original structure or transformed to a more toxic metabolite.[7]
The GIT microbiome can also influence xenobiotic metabolism by the liver, including the expression of some phase 1 (Cyp3a11 and Cyp2c29) and phase 2 hepatic enzymes (glutathione transferase, glutathione peroxidase, acetyltransferases and sulfotransferases),[4] demonstrating the systemic influence of the GIT microbiome.
A range of direct and indirect mechanisms are utilised by the GIT microbiome in the biotransformation of such substances.[3-6] The primary consequence of these direct mechanisms is modification of xenobiotic or metabolite activity via structural biotransformation, while indirect mechanisms results in synergistic communication between the host and microbiome, subsequently influencing various pathways to metabolise or transport xenobiotic substances.[8]
These mechanisms include detoxification, binding, synthesis of pathway intermediates and competition between microbial metabolites and xenobiotics for host enzymes.[6,7,9] Consequently, the activity, clearance and toxicity of the original xenobiotic compound can be increased or decreased.[4,6,10]
Underlying these mechanisms, a range of metabolic reactions are carried out by the GIT microbiota to facilitate xenobiotic metabolism including reduction, hydrolysis, acetylation/deacetylation, demethylation, proteolysis, removal of succinate groups and breaking of amine oxide bonds.[10] The primary enzymes involved are azoreductases, nitroreductases, beta-glucuronidases, sulfatases and beta-lyases as expressed by various members of the microbiome.[7,11]
The specific mechanisms and reactions used to metabolise different xenobiotics is influenced by many factors, two significant ones being the chemical structure and molecular weight of the substance.[8] Examples of how different xenobiotic substances are metabolised demonstrates some of these mechanisms and the overall complexity of this process.
Direct detoxification of xenobiotics and subsequent activation has been observed in the metabolism of sulfasalazine (5-aminosalicylic acid bound to sulfapyridine molecule) by azoreductase enzymes and several other xenobiotics by beta-glucuronidase enzymes.
Azoreductases facilitate the conversion of sulfasalazine to the active form by cleaving the double bond to release 5-aminosalicylic acid.[6,11] Beta-glucuronidase enzyme activity is expressed by many species within the GIT microbiome including proteobacteria, firmicutes and actinobacteria phyla and they have the capacity to increase xenobiotic toxicity via reactivation (e.g. NSAIDs, heterocyclic amines, irinotecan).[8,9] Unsurprisingly, significant variability in beta-glucuronidase enzyme activity between individuals has been observed, contributing further to differences in xenobiotic metabolism across the population.[4]
Conversely the GIT microbiota can directly detoxify substances resulting in reduced xenobiotic activity. Oxalate from plants binds to calcium and other metals and can contribute to renal and cardiac conduction issues. Its metabolism to a less toxic form (oxalyl-CoA) involves the activity of several microbial-derived enzymes – oxalate:formate anti porter, formyl-CoA transferase and oxalyl-CoA decarboxylase.[6] Digoxin activity is also reduced by Eggerthella lenta from the Actinobacterium phylum, with the resulting dihydrodigoxin metabolite being inactive.[6,10]
An example of the direct binding mechanism performed by GIT microbiota can be seen in the metabolism of levodopa (l-DOPA) and heterocyclic amines from charred meats.[6,9] Helicobacter pylori has been observed to interfere with the activity of l-DOPA via direct binding, absorption and metabolism mechanisms, while heterocyclic amines can also be directly bound by GIT microbiota.[6]
Metabolites derived from the GIT microbiota can also influence xenobiotic metabolism, an example of this being the metabolism of acetaminophen.[6]
Following hepatic metabolism of acetaminophen by phase 2 sulfotransferase enzymes (SULT1A1), three metabolites are formed, one of which is N-acetyl-p-benzoquinone imine (NAPQI).[8,12] Increased levels of NAPQI can have detrimental effects on the liver however this varies between individuals partly due to differences in GIT microbial activity.[4,6]
p-cresol is derived from tyrosine and phenylalanine via GIT microbiota metabolism by members of the firmicutes, bacteroidetes, actinobacteria and fusobacterium phyla. Both p-cresol and NAPQI are competitive substrates for SULT1A1, consequently, higher levels of p-cresol can result in higher NAPQI levels due to reduced metabolism of acetaminophen, in part due to the activity of the GIT microbiota.[6,8]
Variability in GIT microbiome composition influences its functional capacity, as observed in a recent study that assessed the differences in xenobiotic metabolising enzyme activity according to age and ethnicity.[13] In a comparative analysis, metagenomics data from eight different countries was used to assess 32 xenobiotic metabolising pathways from six age groups to determine inter-individual, geographic and inter-age variations of GIT microbiota metabolising capacities and associated microbial community compositions.
Overall the results demonstrated that xenobiotic metabolising enzyme abundance and diversity increased with age (0-10 year age group compared with 10-60 year age groups) indicating the impact of increased exposure to xenobiotics with age. Differences in bacterial enzyme composition was also observed to occur between Asian versus American and European subjects, demonstrating potential geographic and genetic influences on the GIT microbiota.
Just as the GIT microbiome significantly influences xenobiotic metabolism, exposure to xenobiotics can induce structural, compositional and functional changes to the GIT microbiome, their downstream metabolites and gene expression, consequently influencing xenobiotic metabolism.[2,7,14]
A range of xenobiotics have been observed to alter GIT microbiota composition, structure and abundance, including air pollutants, pesticides (glyphosate), insecticides (chlorpyrifos), polychlorinated biphenyls, organophosphates, heavy metals (mercury, cadmium, lead, arsenic) and medications (antibiotics, proton pump inhibitors)[2,14,15] and dietary components (artificial sweeteners, emulsifying agents).[8]
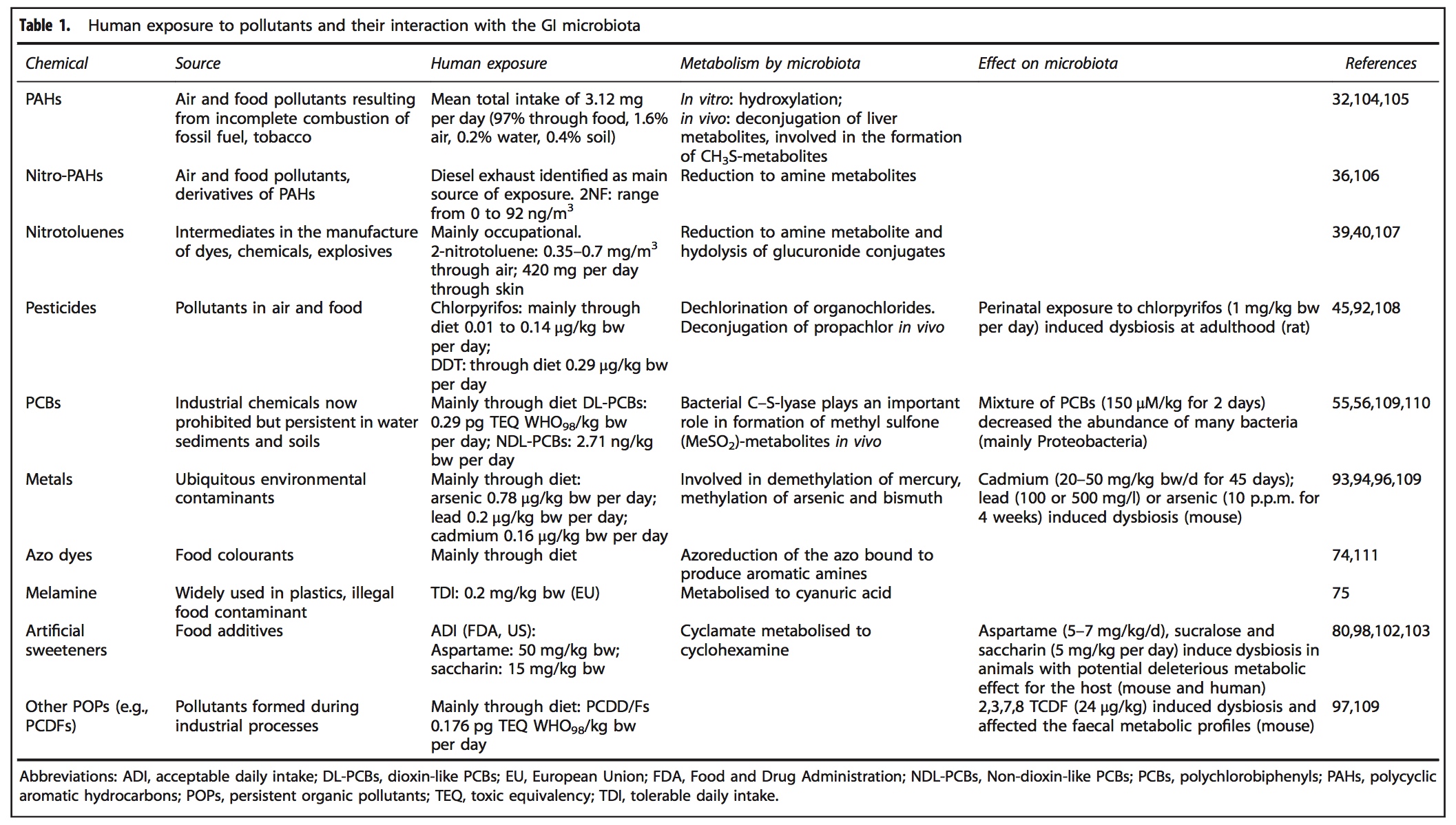
Image Source [7]
Overall, what is currently known about xenobiotic metabolism highlights the importance of taking into account the significant variability occurring between individuals in relation to xenobiotic exposure (frequency, dose and the time period it occurs over) and the capacity to metabolise them as a result of genetic, compositional and functional differences in hepatic and GIT microbiome activity. The health of the GIT microbiota is an essential consideration in maximising the body’s capacity to effectively metabolise exogenous and endogenous xenobiotics both locally and systemically.
As with many aspects of the GIT microbiome, much is yet to be ascertained, including the influence of inter-individual differences in microbial species, enzymatic and metabolite composition and functionality on xenobiotic metabolism, and how to manipulate the GIT microbiome to facilitate the effective biotransformation of the many xenobiotics we are continually exposed to in our modern world.[8,10]
References
- Jimenez-Chillaron JC, Nijland M, Ascensao AA, et al. Back to the future: transgenerational transmission of xenobiotic-induced epigenetic remodelling. Epigenetics 2015;10(4):259-273. [Full text]
- Lu K, Mahbub R, Fox JG. Xenobiotics: interaction with the intestinal microflora. ILAR J 2015;56(2):218-227. [Full text]
- Kim DH. Gut microbiota-mediated drug-antibiotic interactions. Drug Metab Dispos 2015;43(10):1581-1589. [Full text]
- Snedeker SM, Hay AG. Do interactions between gut ecology and environmental chemicals contribute to obesity and diabetes? Environ Health Perspective 2012;120(3):332-339. [Full text]
- Jandhyala SM, Talukdar R, Subramanyam C, et al. Role of the normal gut microbiota. World J Gastrenterol 2015;21(29):8787. [Full text]
- Carmody RN, Turnbaugh PJ. Host-microbial interactions in the metabolism of therapeutic and diet-derived xenobiotics. J Clin Invest 2014;124(10):4173-4181. [Full text]
- Claus SP, Guillou H, Ellero-Simatos E. The gut microbiota: a major player in the toxicity of environmental pollutants. Biofilms and Microbiomes 2016;2:Article No.16003. [Full text]
- Spanogiannopoulos P, Bess EN, Carmody R, et al. The microbial pharmacists within us: a metagenomic view of xenobiotic metabolism. Nat Rev Microbiol 2016;14(5):273-287. [Full text]
- Swanson HI. Drug metabolism by the host and gut microbiota: a partnership or rivalry? Drug Metab Dispos 2015;43(10):1499-1504. [Full text]
- Nayak RR, Turnbaugh PJ. Mirror, mirror on the wall: which microbiomes will help heal them all? BMC Medicine 2016;14:72. [Full text]
- Haiser HJ, Turnbaugh PJ. Developing a metagenomics view of xenobiotic metabolism. Pharmacol Res 2013;69(1):21-31. [Full text]
- Guidry A, Falany C. Identifying the structure-function activity of human cytosolic sulfotransferase (SULT) 1C4. FASEB J 2015;29(1):Supp 662.6. [Abstract]
- Das A, Srinivasan M, Ghosh TS, et al. Xenobiotic metabolism and gut microbiomes. PLoS One 2016;11(10):e0163099. [Full text]
- Maurice CF, Haiser HJ, Turnbaugh PJ. Xenobiotics shape the physiology and gene expression of the active human gut microbiome. Cell 2013;152(1-2):39-50. [Full text]
- Velmurugan G, Ramprasath T, Swaminathan K, et al. Gut microbial degradation of organophosphate insecticides-induces glucose intolerance via gluconeogenesis. Genome Biol 2017;18(1):8. [Full text]
DISCLAIMER:
The information provided on FX Medicine is for educational and informational purposes only. The information provided on this site is not, nor is it intended to be, a substitute for professional advice or care. Please seek the advice of a qualified health care professional in the event something you have read here raises questions or concerns regarding your health.





